Virus Isolation ‐ genome detection
Detection of IBV in clinical samples can be done using real time RT‐PCR. Sequencing the spike glycoprotein gene is routinely done to identify different IBV types. Identification of more than one type of IBV in a single sample can be done using RT‐PCR and RFLP analysis.s Bronchitis (IB) is a viral disease affecting chickens of all ages.
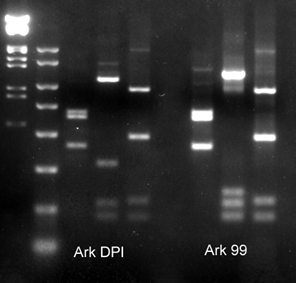
IBV typing by RT‐PCR and RFLP
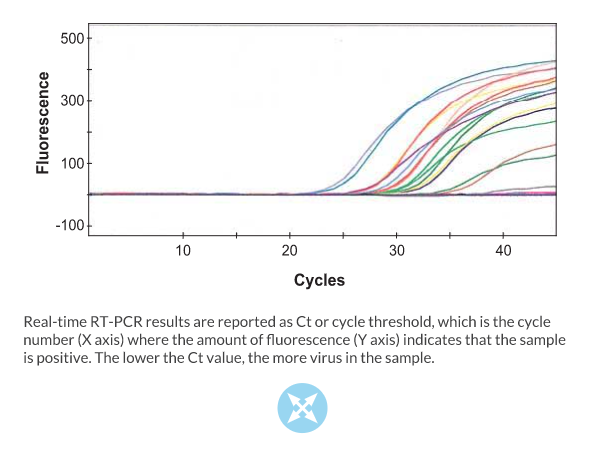
Real‐time RT‐PCR is a typical RT‐PCR reaction that includes a fluorescent dye or labeled probe that is used to detect the amplified DNA as it is being synthesized during the reaction. The advantage of real‐time RT‐PCR is that it can be conducted in a matter of minutes and it is quantitative. Real‐time RT‐PCR has been developed for detection of IBV (Callison et al. J. Virol. Methods 138:60‐65, 2006). This highly sensitive method is generally used to screen a large numbers of samples for the virus. This particular method does not identify IBV type.
IBV typing by nucleic acid sequencing
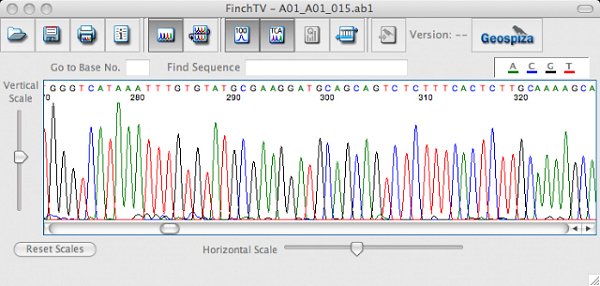
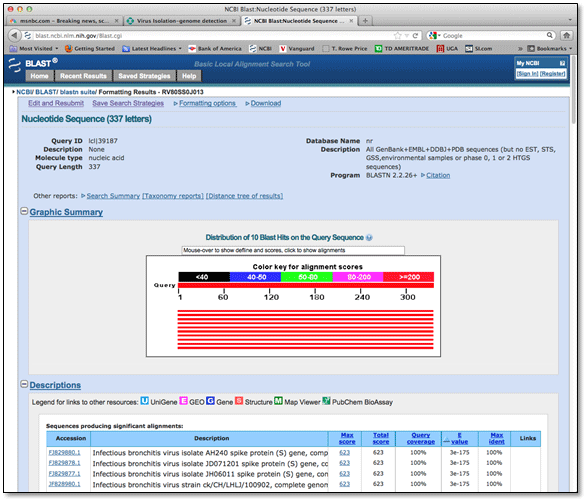
Molecular typing of IBV is routinely conducted by sequence analysis of the spike glycoprotein gene. Typically the hypervariable regions of the spike gene, which are highly variable sequence regions that correlate with IBV serotype, are RT‐PCR amplified. The amplified product can be directly sequenced to determine the type of IBV in the sample. In addition, a BLAST or Basic Local Alignment Search Tool, can be used to find similar sequences in GenBank (http://www.ncbi.nlm.nih.gov/), a database of nucleic acid sequences from all over the world. Phylogenetic analysis conducted with the viruses in the database will show how closely the viruses are related. See the Phylogenetic tree.
IBV typing by real‐time RT‐PCR
The spike glycoprotein of IBV contains unique sequences that can be used to identify different types of the virus. First RT‐PCR is conducted to amplify the spike gene then restriction enzymes that act like molecular scissors, specifically cut the amplified product into unique lengths called restriction fragments. When visualized on an agarose gel, the restriction fragments form a pattern that can be used to identify the type of IBV in the sample. This technique can identify all known serotypes and variant viruses of IBV. More importantly, it can be used to identify more than one virus type in a single sample.